Cell Motility and the Cytoskeleton –
Microfilaments CS3
Cells display an amazing variety of
movement. They swim, crawl, contract,
extend and move things around inside.
These movements perform an equal variety of functions such as to capture
prey or invading organisms, to feed, or communicate with other cells. All this is accomplished by the
microfilament and/or microtubule systems in association with various “motor
proteins” which are capable of transforming chemical energy in the form of
nucleotides into kinetic energy.
The Myosins
The myosin are a diverse group of motor proteins which interact with microfilaments to produce force. What defines a myosin is a conserved “head” domain which binds actin and hydrolyses Mg-ATP. This head domain is the motor. A generalised scheme of the actin-myosin cycle is outlined below. There are at least 11 classes of myosins but this number is likely to be very much greater. These have been identified by cDNA technology (largely RT-PCR), and very scant biochemical details have been gleaned from these different classes with the exception of myosin II and myosin I. Fortunately these same classes represent the most abundant forms and possibly the two most divergent.
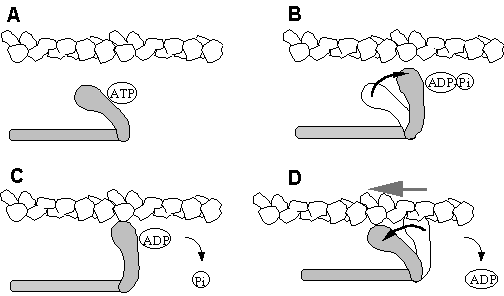
Figure
12. The Myosin Crossbridge Cycle. A.
ATP binding to a cleft at the “back” of the head causes a conformation
which cannot bind actin. B. As the ATP is hydrolysed, the head swings
back about 5nm to the “cocked” position the ADP and Pi remain
bound. C+D. The force generating stages.
When the Pi leaves the myosin, the head binds the actin and the “power
stroke” is released as the head bind actin.
ADP is released to continue the cycle. At this stage the head in bound
to actin in the “rigor” or tightly bound state.
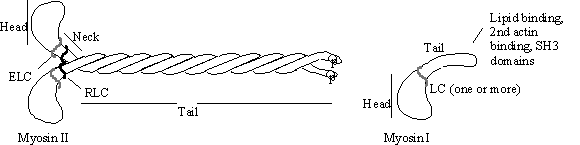
Figure
13. Myosin II has a long coiled coil - tail which
polymerises with other molecules to give large bi-polar filaments (figure 13)
in muscle and non-muscle cells. The
assembly state is regulated by phosphorylation close to the tail end, whereas
the contractile activity is controlled by phosphorylation of the regulatory
light chain (RLC). Myosin I also has
light chains, some of which are calmodulin-like or calmodulin itself. The tails of myosin Is are extremely variable,
some bind phospholipid, actin and some contain poly-proline binding SH3
domains.
Myosin I type motors have various lengths of “tails” associated. Some of these bind phospholipid, others have SH3 domains. It is generally held that the function of myosin I motors is to move vesicles around the cell. but other functions have been ascribed. The microvillus has a myosin I whose function is assumed to be to keep the membrane from “riding up” with wear! In Acanthamoeba one iso-form (myosin 1c) has been shown to be essential for contractile vacuole function while others have been shown to be involved in pinocytosis. It is much easier to describe the function of myosin II, in muscle at least:-

Figure
14. The sarcomere in a semi-contracted state. As myosin is activated this arrangement
causes the Z-discs to move towards each other.
The troponin complex senses calcium and forces tropomyosin to move
position on the filament which controls access to the myosin heads. Myosin thick filaments are arranged as
anti-parallel bundles each with a central bare zone. These thick filaments are associated with “protein C” which
presumably helps keep the structure together.
Thick filaments are held to the Z-disc by Titin, a giant elastic
protein. Thin filaments are stabilised
by nebulin, and held to the Z-disc by a-actinin and capZ.
Microfilament dynamics and cell locomotion.
In order to induce actin polymerisation in cells
one requires to add a nucleation site.
This has been demonstrated by the microinjection of stable actin
nuclei. Recently, it has come to light
that certain are capable of using the cellular actin to propel themselves
through the cytoplasm of one cell and into neighbouring cells. The speed at which the bacteria are driven
is cell specific varying between 0.02 and 1.5µm/sec. Propulsion is thought to be achieved by the production of an
actin nucleating activity expressed on the surface of the bacteria. The mechanism is not yet understood, but
appears to involve cellular components such as profilin. Listeria-induced actin
polymerization may be considered as a simplified leading lamella and this
system promises to be a useful tool to unravel the role of nucleation in cell
locomotion.
Polymerization
occurs at the leading edge by some presently unknown nucleator;
depolymerisation probably takes place everywhere but the leading edge. Filaments polymerising at the intracellular
face of the plasma membrane are oriented with their pointed ends facing the
nucleus. As a result of the different assembly
rates at the two ends, monomers are likely to add at the barbed end and travel
down the filament to fall off at the pointed end, a phenomenon termed
"treadmilling" (Figure 10, page 7 CS2). Treadmilling has been postulated to drive lamellae forwards, but
the rate of treadmilling is limited by the rate of depolymerisation, which at
0.8 monomers per second would permit an extension of 0.13µm per minute. This is
much too slow to account for the rapid rates of locomotion achieved by many
cell types, but perhaps the answer is severing proteins.
Theoretically
at least, it is possible that the energy available from the polymerization of actin could be used to produce
lamellar protrusions. Certainly it has
been found that pure unpolymerised actin within membrane vesicles is able to
distort the membrane when the actin polymerises. Recently a model for how actin polymerization alone might drive
filopodial protrusion has been presented.
The "Brownian Ratchet" model states that a protrusive force is
exerted during polymerization of actin by utilising the energy of Brownian
motion in which the plasma-membrane is permitted to diffuse in only one
direction by the growing actin filament (Figure 15). The maximum force produced by a single actin filament according
to this model is 9 pN (six times that produced by a single myosin head!). A striking feature of this model is that it
requires the barbed end of the filaments to be free, i.e. not capped by other
proteins. Consequently the Brownian
ratchet model is not easy to reconcile with mechanisms outlined in the
preceding paragraph, which describe the importance of nucleated assembly in
cell locomotion. Perhaps we can fuse
the Brownian ratchet model and the nucleation release models by assuming that
other proteins localised at the leading edge of locomoting cells provide
"privileged conditions" for the assembly of actin. Such conditions might be provided by
activation of the PIP2 cycle.
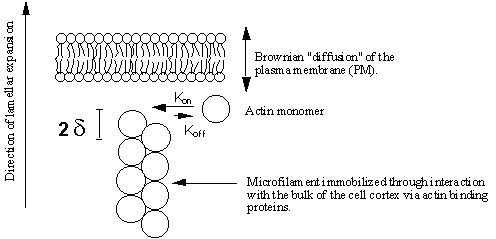

Figure 15 The Brownian ratchet model of lamellar protrusion. According to this hypothesis, the distance
between the plasma-membrane (PM) and the filament end fluctuates randomly. At a
point in time when the PM is most distant from the filament end, a new monomer
is able to add on. Consequently, the PM
is no longer able to return to its former position since the filament is now
that much longer. The filament cannot
be pushed backwards by the returning PM as it is locked into the mass of the
cell cortex by actin binding proteins.
In this way, the PM is permitted to diffuse only in an outward
direction. The maximum force which a
single filament may exert is related to the thermal energy of the actin monomer
by kinetic theory according to the above equation.
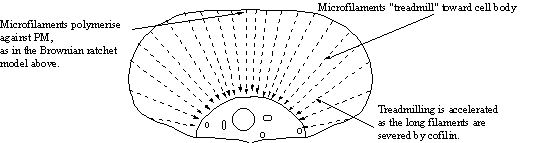
The Brownian ratchet together with accelerated “Treadmilling” may explain lamellar protrusion. Actin is polymerised at the leading edge and pushes the PM forward as above. Actin continues to treadmill down the filaments which depolymerise rapidly toward the cell body due to the action of filament severing proteins possibly belonging to the cofilin family.
The Role of Myosins in Cell Motility and Locomotion
Dictyostelium provides an excellent test for myosin function as this
organism is haploid (one genome copy per cell) it is comparatively easy to
knock out gene function by homologous recombination. Knocking out the single gene encoding the myosin II heavy chain
produced cells that were devoid of myosin II and established that these cells
were still able to move (at reduced rates) but could not carry out cytokinesis
in the normal fashion. These
experiments indicate that myosin II is not absolutely required for the
locomotion of Dictyostelium, but
possibly "fine tunes" the process.
In addition to the one myosin II gene, Dictyostelium expresses two myosin XI genes and at least six myosin
I genes. So far 4 of the myosin I genes
(named myoA - myoD) have been knocked out, these cells seem healthy in the lab
at least but are impaired in some aspects of motility. These knockout reveals that myoA and myoB
mutants locomoted somewhat slower than wild type but that myoC and myoD mutant
were as fast. MyoB and myoC were
impaired in phagocytosis and all were impaired in pinocytosis. Thus there seems to be some overlap in the
function of the myosin I family, but much more work is needed to establish why
the cell expresses so many types.
The Transduction of Extracellular Motility Signals.
Extracellular signals of a chemical nature are
generally received at the cell surface by binding a specific receptor, usually
a transmembrane protein. Binding of
this extracellular signal induces a change in the intracellular domain of the
receptor which affects the properties associated with the receptor, such as
kinase activity or G-protein binding.
The signal transduction cascade produces a series of intracellular
messengers such as calcium release, polyphosphoinositide hydrolysis or protein
phosphorylation.
It
has often been observed that cells respond to growth factors by increased
motility, both of stimulated locomotion and ruffling. Growth factors mediate
their effects through a variety of different intracellular signalling pathways
and these have been suggested to modulate motility differentially. For example, growth factors that utilise the
phosphoinositide pathway are found to increase motility while those that
increase cAMP levels depress motility and severely alter cell shape and
cytoskeletal organization. Clues are
beginning to emerge concerning mechanisms connecting extracellular signals to
the actin based cytoskeleton. Two small
GTP-binding proteins are known to be involved in modulating actin
dynamics. One of these, rho, regulates the assembly of focal
contacts (this topic will be covered later) and stress fibres in response to
various growth factors, and another, rac,
regulates growth factor stimulated membrane ruffling. The integrins are a group of integral membrane proteins that bind
both to the extracellular matrix on the outside of the cell and to cytoskeletal
components such as a-actinin and talin on the cytoplasmic side of the plasma
membrane. Clustering of integrins at
focal contacts (or artificially by antibodies) results in the phosphorylation
of a host of cytoskeletal proteins on tyrosine residues, concomitant with actin
polymerization.

References:-
Signal Transduction and actin Zigmond “Signal transduction and
actin filament organization”
Current Opinion in Cell Biology 8, 66-73. March 1996
Listeria motility Lasa
& Cossart “Actin-based bacterial motility”. Trends in Cell Biology
March
1996, 6, 109-114.
Cell locomotion Mitchison & Cramer
“Actin-based cell motility and cell locomotion”
Cell
84 371-379. Feb, 1996
Lauffenburger
& Horwitz “Cell migration; a physically integrated molecular process.
Cell
84 359-369. Feb, 1996
Myosin I and motility Ostap & Pollard. “Overlapping functions
of myosin-1 isoforms?” J.Cell Biol. 133,
221.
April
1996
Paul McLaughlin (ICMB) has produced an excellent interactive
tutorial on Actin & Myosin Structures see:-
http://chon.bch.ed.ac.uk/paul/TEACHING/HONS/MUSCLE/
Myosin head structure is available on the
WWW:- http://salus.med.uvm.edu/~guilford/myosin.html
Please direct any questions to me at:-
Room 444 or lab 446 fourth floor HRB. Tel (0131) 650 3714 or 3712. E-mail SKM@srv4.med.ed.ac.uk